Detailed Running
This section provides a detailed explanation of how to run the QLIMR module, either using a Docker container or locally on the user’s machine. It also includes information about the module’s general structure, including its dependencies, components, and the algorithm implemented in the C++ source code for computing neutron star observables.
Docker Container
As highlighted in the Quick Start section, the easiest way to run QLIMR is via a Docker container. This method is preferred because users may be working with various operating systems, and manually installing the necessary libraries can be complex and error-prone. Docker simplifies this by offering a consistent environment that works across all systems. In this section, we outline two methods for running QLIMR in a Docker container: the first method involves pulling an image from the GitLab registry, while the second method entails cloning the entire repository from GitLab, building, and running the container.
Method 1: Pulling image
This subsection expands on the Quick Start section, offering detailed instructions on how to run the module using Docker by pulling the QLIMR image from the GitLab registry.
Installation. Docker is a platform that allows applications to run in isolated environments, known as containers. To use Docker, it must first be installed on the user’s machine, similar to how other software is installed. Detailed instructions for installing Docker on various operating systems (Linux, macOS, Windows) can be found in the official Docker documentation: Docker Installation Guide.
Login to GitLab Container Registry. To begin, Docker Desktop should be open and running. Once Docker is active, the user needs to log in to the GitLab container registry to access the Docker image required for the QLIMR module.
A Docker image is a lightweight, stand-alone, executable package that contains everything necessary to run a piece of software, including the code, runtime, libraries, and system tools. The GitLab container registry serves as a centralized repository where these images are stored, managed, and shared, simplifying the process of deploying software. The Docker image for the QLIMR module is hosted within this registry, while the module’s source code resides in the corresponding GitLab repository.
To log in, the user must have a GitLab account. GitLab accounts are free and provide access to both public and private container images.
Steps to Log in
The user should open the terminal.
Run the following command to log in to the GitLab container registry:
docker login registry.gitlab.com
When prompted, the user will need to enter their GitLab username and password. If two-factor authentication (2FA) is enabled, a Personal Access Token (PAT) should be used instead of the password.
After entering the credentials, the terminal will display the message: “Login Succeeded”.
Once logged in, the user will have access to the required image for the QLIMR module from the GitLab container registry.
Pull the Docker Image. Once Docker Desktop is open and the user has logged into the GitLab container registry, the next step is to pull the QLIMR Docker image. Pulling a Docker image means downloading it from the container registry to the local machine so that it can be used in Docker.
The QLIMR Docker image is stored in the GitLab container registry under the following address: registry.gitlab.com/nsf-muses/ns-qlimr/qlimr:v1.0.0 The name of the Docker image is called qlimr and the tag v1.0.0 refers to the first stable version of the QLIMR module. Using version tags is important for ensuring compatibility and stability, as newer versions of the module may include updates or changes that could affect functionality.
To pull the QLIMR Docker image, the user should execute in a terminal the following command:
docker pull registry.gitlab.com/nsf-muses/ns-qlimr/qlimr:v1.0.0
This command will instruct Docker to download the image from the GitLab container registry. Once the image is pulled, it will be available for use locally in the Docker environment. If a different version of the module is needed in the future, the user can simply replace v1.0.0 with the desired version tag, such as v1.1.0, latest, or any other available tag corresponding to a specific version. The user can verify that the image was successfully downloaded an is available for use in Docker Desktop.
Directories setup. The user should create a main directory named qlimr in their preferred location, along with two subfolders: input for the necessary input files to run the module, and output for storing the results, where the observables generated after running the module will be saved. Once the directories are created, the user should navigate into the qlimr directory by executing the following command:
mkdir qlimr && mkdir qlimr/input qlimr/output && cd qlimr
Add Equation of State data file. The user should place the Equation of State (EoS) data file in the input folder. QLIMR supports two formats for the EoS data file: MUSES EoS tables or the QLIMR internal format, where the first column represents the total energy density and the second column represents the pressure. The columns must be delimited by a comma (“,”) and the file should be saved in either .csv or .h5 format. Additionally, the file must be named as
eos. This fixed name is important, as the QLIMR module will not recognize the EoS file otherwise. The naming convention is required by the project’s cyberinfrastructure. However, despite this fixed name, the user can specify the EoS to be used in the input parameters before running the module.
Execution. To run the QLIMR module with Docker, the user should execute the following command:
docker run --platform linux/amd64 -it --rm --name qlimr \ -v "${PWD}/input:/opt/input" \ -v "${PWD}/output:/opt/output" \ registry.gitlab.com/nsf-muses/ns-qlimr/qlimr:v1.0.0 python3 qlimr.py --eos_name EoS
Breaking Down the Command:
`docker run`:
This tells Docker to run a container using the specified image.
`–platform linux/amd64`:
This flag ensures the Docker container runs with the linux/amd64 architecture, making it compatible with most Linux systems and ensuring consistent behavior. Note: If the user is running Docker on a different platform (e.g., macOS or Windows), Docker will automatically handle cross-platform compatibility. In some cases, it may not be necessary to specify the platform, but it’s included here to ensure the correct architecture is used for consistent execution.
`-it`:
The -i option makes the Docker container run in interactive mode (keeps the input stream open). The -t option allocates a pseudo-TTY (terminal) to the container, enabling interactive use, such as viewing output in the terminal.
`–rm`:
This flag removes the container once it finishes running. It helps keep the system clean by automatically deleting the container after execution, avoiding the accumulation of unused containers.
`–name qlimr`:
This assigns the name qlimr to the running container. Naming the container can help the user easily reference or manage it later, such as stopping or inspecting the container.
`-v “${PWD}/input:/opt/input”`:
This option mounts the input directory from the user’s local machine (${PWD}/input, where ${PWD} is the current working directory) to the /opt/input directory inside the container. This ensures that the input files, such as the Equation of State (EoS) data file, are accessible to the QLIMR module within the container.
`-v “${PWD}/output:/opt/output”`:
Similarly, this mounts the output directory from the local machine (${PWD}/output) to /opt/output inside the container. This allows the QLIMR module to store the generated results (such as observables) in the local output folder after the execution.
`registry.gitlab.com/nsf-muses/ns-qlimr/qlimr:v1.0.0`:
This specifies the Docker image to use, which is the qlimr image hosted in the GitLab container registry. The image tag v1.0.0 ensures that the user is running a specific version of the QLIMR module (version 1.0.0). If a different version is needed, the tag can be updated accordingly.
`python3 qlimr.py –eos_name EoS`:
This part runs the qlimr.py script using Python 3 inside the container. The –eos_name EoS argument specifies the name of the Equation of State (EoS) file to use. In this case, EoS refers to an example name provided by the user, and the user can choose any name they desire. This is the only required flag to run the module. For additional input parameter options, users can refer to the source code section.
Output. Once the QLIMR module has finished execution, the user can retrieve the output file named
observables.csvfrom the output directory. This file will be in either .csv or .h5 format, depending on the user’s specifications. The observables.csv file contains the results of the module, including the obtained observables. The user can open this file using a text editor, spreadsheet software, or any compatible tool to view and analyze the results.
Method 2: Cloning QLIMR repository
The alternative method for running QLIMR in a container involves cloning the entire repository from GitLab, followed by building and running the Docker container. Below are the steps to use this method:
1. Installation. As with the previous method, Docker must be installed in order to run QLIMR in a Docker container. Follow the Docker Installation Guide to install Docker based on your operating system.
2. Open Docker Desktop to prepare Docker for use on your machine. If you are using macOS with M1 or M2 chips, it is recommended to go to the Settings menu in the top right corner of Docker Desktop and enable the option Use Rosetta for x86/amd64 emulation on Apple Silicon. This will make Docker execution much more efficient.
3. Clone the module’s repository: Open the QLIMR repository at https://gitlab.com/nsf-muses/ns-qlimr/qlimr. In the upper right corner of the page, click on “Code” and copy the URL under “Clone with HTTPS.” Then, open a terminal on your machine in your preferred directory and run the following command, replacing <URL> with the copied URL, as shown below:
git clone https://gitlab.com/nsf-muses/ns-qlimr/qlimr.git4. Provide Equation of State. The user needs to provide an Equation of State (EoS) and prepare the input file required to run the module. An example EoS file, named
Sly_muses_format.csv, is located in the test folder of the QLIMR repository. For more information about this example, refer to the Example section. The user should copy this file into the input folder and rename it aseos.csv. This can be done in the terminal by running the following command:cp test/Sly_muses_format.csv input/eos.csv5. Build the QLIMR image. Navigate to the folder where you cloned the repository and ensure you are in the same directory as the Dockerfile. The Dockerfile contains the necessary instructions to configure, manipulate, and install components inside the container. To build the QLIMR image, execute the following command:
docker build -t qlimr:latest .If you are using a machine with an Apple Silicon chip, it’s better to include the platform option to ensure compatibility.
docker build --platform linux/amd64 -t qlimr:latest .
Run the container. To run the container, execute the following command:
docker run --platform linux/amd64 -it --rm --name qlimr \ -v "${PWD}/input:/opt/input" -v "${PWD}/output:/opt/output" \ qlimr:latest python3 qlimr.py --eos_name EoS \ --compute_inertia 1 \ --compute_love 1 \ --compute_quadrupole 1 \ --compute_mass_and_radius_correction 1 \ --local_functions 1The components of this command were explained in the previous Docker method. In this case, additional input flag options are included to compute more observables based on the Sly EoS (see the Example section for details).
7. Retrieve output result. After running the container, the user will see in the terminal that the execution has completed and all the macroscopic observables have been calculated. To retrieve the results, the user should navigate to the output folder inside the repository. There, they will find the
observables.csvfile, which contains all the requested observables. If the user also requested the local functions for the entire neutron star sequence, multiple files will appear, each labeled with the corresponding central energy density.
Local Usage & YAML files
This subsection outlines the steps to run QLIMR locally on the user’s machine without using a Docker container. This method is ideal for users who want to modify and test the QLIMR source code for their specific use cases. There are two ways to run it locally: i) by using the Python wrapper, which handles the internal workflow of the module for preprocessing and postprocessing data, or ii) by generating the input configuration file required to run QLIMR. For more details about the code structure, please refer to the Code Structure section.
Method 1: Using python wrapper
The following set of instructions builds upon those provided in the Quick Start section of this documentation, offering a more detailed explanation of each step. The general structure remains the same. The steps are as follows:
Install dependencies. The first step before running QLIMR locally is to install and set up all the required dependencies. There are two types of libraries needed:
Python libraries: numpy, pandas, PyYAML, openapi-core, and muses_porter
C++ libraries: yaml-cpp and the GNU Scientific Library (GSL)
For more details about these libraries and dependencies, please refer to the Code Structure subsection.
Clone the repository: Repeat step 3 from Method 2 in the Docker Container section.
Provide Equation of State: Repeat step 4 from Method 2 in the Docker Container section.
Build qlimr executable: Navigate to the source code folder, named src, and build the qlimr executable by running
cd src/ && makeThe make tool automates the process of compiling and linking the C++ source code. It reads the Makefile, which contains the necessary instructions for compiling the .cpp files and linking them with the required libraries (such as those installed previously). The Makefile specifies how to handle dependencies, ensuring that the .hpp header files and corresponding .cpp files are correctly compiled. Once the executable qlimr is built, it can be run multiple times with different parameter configurations, without needing to rebuild the code. This process of “building” refers to creating the executable, which is necessary before QLIMR can be executed.
Execution. To execute QLIMR locally using the python wrapper, use the following command
python3 qlimr.py --eos_name EoSThe
qlimr.pyscript serves as the QLIMR wrapper, handling the execution of the application, including data preprocessing and postprocessing. It parses the input EoS data file, extracting the energy density and pressure columns. Additionally, it generates the necessary YAML configuration files required to run the module. The postprocessing step primarily involves formatting the output into either.csvor.h5files.Output. Retrieve the output file named
observables.csvfrom the output directory that is already in the repository.
Method 2: Creating YAML files and using the Makefile
Follow steps 1 and 2 from the previous method.
Provide Equation of State: The user must provide an Equation of State (EoS) file to run QLIMR. As mentioned earlier in the documentation, there are two conventions for the columns in the EoS file: 1) the MUSES EoS tables, and 2) the QLIMR format, where the first column represents total energy density and the second column represents pressure. The columns should be delimited by a comma (“,”) and the file must be in either .csv or .h5 format. Additionally, the file should be named
eosfor the MUSES table convention.If the user provides the MUSES convention format (as detailed in the Parameters section), the file must be parsed using a script called
preprocess.py. This script extracts the energy density and pressure columns, creating a new EoS file calledeos_validated.csv. Ultimately, QLIMR only uses the total energy density and pressure in a two-column format. To parse the EoS data required by QLIMR, run the following command inside the src folder:python3 preprocess.pyIf the user already has a file in this format, they can simply rename it to eos_validated.csv and skip this step.
Create configuration file: The user can generate the input configuration file based on the input flag parameters explained in the Parameters section. This can be done by running the create_config.py script located in the /src folder. The script accepts input parameter flags to customize the configuration. To create a basic configuration YAML file for QLIMR, execute the following command:
python3 create_config.py --eos_name EOSOnce the previous command is executed, a new YAML file named config.yaml is generated inside the input folder. The generated
config.yamlfile will look as follows:inputs: eos_name: EOS options: {} outputs: {}Validate the configuration file: The generated configuration file may contain incomplete or invalid input parameters. For example, the user might accidentally provide a string where a numeric value (e.g., a double) is expected. To avoid such issues, a validation process is available to ensure that the provided data is correct and to complete any missing input parameters required by the module. This can be done by executing the following script which is located inside the src folder:
python3 Validator.pyOnce the validator script is executed, a new configuration file named
config_validated.yamlis generated inside the input folder of the repository. This file serves as the final configuration, which the C++ code will read in order to execute QLIMR. Below is an example of how the config_validated.yaml file will look:inputs: A22_int: 1.0 R_start: 0.0004 eos_name: EOS final_epsilon: .inf initial_epsilon: 250.0 resolution_in_NS_M: 0.05 single_epsilon: 700.0 wb11_c: 0.1 options: eps_sequence: true output_format: csv stable_branch: true outputs: compute_all: false compute_inertia: false compute_love: false compute_mass_and_radius_correction: false compute_quadrupole: false local_functions: falseNote: The user can use this YAML file as a reference to manually create the input configuration parameters for the module, should they prefer not to run the validator. However, it is the user’s responsibility to ensure the validity of the input data.
Build QLIMR executable: To build the C++ source code, a Makefile is used to manage the compilation and linking process, incorporating all the required libraries. Before building the executable, open the Makefile located inside the /src folder and ensure that the paths to all libraries are correct. For example, on macOS with M1 chips, the paths to the required libraries can be updated by modifying the following variables to the correct paths:
#------------------------- Local flags on M1 -------------------------------- CXXFLAGS = -I/opt/homebrew/Cellar/gsl/2.8/include -I/opt/homebrew/Cellar/yaml-cpp/0.8.0/include -I/opt/homebrew/Cellar/libomp/18.1.8/include -g3 -Xclang -fopenmp -Wall -Wextra -Wpedantic -Wshadow -Wconversion -Wsign-conversion -Werror LDFLAGS = -L/opt/homebrew/Cellar/gsl/2.8/lib -L/opt/homebrew/Cellar/yaml-cpp/0.8.0/lib -L/opt/homebrew/Cellar/libomp/18.1.8/lib -g3 -Xclang -fopenmp STD = -std=c++11 LIBS = -lgsl -lgslcblas -lm -lyaml-cpp -lomp OPTIMIZATION = -O3 RM_RF = rm -rfNext, execute the following command to build and run QLIMR:
make && make runIt’s worth noting that the user typically only needs to build QLIMR once, unless they intend to modify the source code to achieve specific results.
Output. Retrieve the output file named
observables.csvfrom the output directory that is already in the repository.
Code Structure
This section provides a clear overview of the module’s structure, focusing on its main parts and how they work together. It includes three key subsections: Dependencies, which lists the necessary libraries and tools; Components, which describes the important pieces of the module; and Source Code, which explains how the moduele’s physics calculations are implemented.
The module’s source code is written in C++, supported by two Python layers—one for preparing the data and
another for postprocessing. The entire application is wrapped in a Python script called qlimr.py, where
users can input their parameters as flag arguments. This script can be executed on a local machine or
within a Docker container.
In the Source Code subsection, detailed information about the implementation, including inputs and outputs is provided to describe how the module operates.
Dependencies
A description of each required library for the Python layers and the C++ source code is provided. If the module is run through Docker, these libraries are included in the Docker image. For users planning to execute the module on a local machine, please refer to the Execution section for detailed information on local installation.
Python Libraries
NumPy is a powerful library for Python that simplifies working with numerical data. It provides support for large, multi-dimensional arrays and matrices, along with a collection of mathematical functions to perform operations on these arrays efficiently.
pandas is a versatile library for Python that makes working with structured data easy. It provides powerful data structures, like DataFrames, which are similar to tables in a spreadsheet or SQL database.
PyYAML is a Python library that makes it easy to work with YAML, a human-readable data serialization format. It allows you to easily parse YAML files into Python objects and convert Python objects back into YAML format.
openapi-core is a Python library that simplifies working with OpenAPI specifications. It allows you to easily validate and interact with API requests and responses based on OpenAPI definitions.
muses_porter Is a Python library developed for the MUSES Collaboration cyberinfrastructure to handle .csv and .hdf5 formats and column conventions.
C++ Libraries
yaml-cpp is a human-readable data serialization standard that can be used in conjunction with all programming languages and is often used to write configuration files. The C++ source code uses .yaml format files for reading the inputs specified by the user and outputs a status.yaml file after execution.
GNU Scientific Library is a versatile mathematical toolkit developed by the GNU Project for C and C++ and provides a wide set of mathematical functions and algorithms. QLIMR integrates the GSL library due to its exceptional performance ensuring efficient and accurate solutions for tasks involving interpolation, numerical integration, and solving ordinary differential equations.
Components
In this section, the basic structure of the code is explained, focusing on its use within a Docker container. This helps avoid
compatibility issues across different operating systems and their dependencies. First, an overview of the Dockerfile will be provided,
which sets up the container with the necessary tools and libraries, including a method to reduce the image size. Next, qlimr.py will
be introduced as the script that the user interacts with, which contains an internal workflow consisting of five main steps: creating
a user configuration file, validating input data, preprocessing the EoS data, executing the C++ code, and postprocessing the results.
DOCKER CONTAINER
A Docker container is a lightweight, portable environment that allows applications to run consistently across different operating systems. By using a container, conflicts with dependencies that may arise from variations in system configurations are avoided.
Inside the module’s repository, there is a file called Dockerfile, which is a script containing instructions to set up the container with
all the necessary dependencies for the module. The Dockerfile follows a multi-stage approach to minimize the size of the final image by
including only what is essential. This method not only reduces the image size but also speeds up the building process. The multi-stage process
includes three main parts:
Note
1. Building qlimr. In this stage, a common MUSES image is pulled, designed for use by all modules and containing basic packages like yaml-cpp and make. The GSL library is then installed, which is required for compiling the source code. Next, the /test and src/ folders are copied into the image. Finally, the QLIMR source code is compiled.
2. Installing Python dependencies. This Dockerfile stage starts by pulling a slim Python 3.11 base image. It references a
requirements.txt file from the module’s repository, which outlines the necessary Python dependencies as detailed in the dependencies
subsection above. To accommodate dependencies hosted on Git, Git is then installed. After updating the package list and installing Git,
the package lists are removed to optimize space. Finally, the requirements.txt file is copied into the image, and the specified Python
dependencies are installed.
3. Final image setup. In this final stage, a slim Python 3.11 base image is used to set up the final image. It installs make, defines a user with a specified UID, and creates a home directory at /opt. The stage then installs previously compiled Python and C++ dependencies from earlier stages, along with the source code, unit tests, API files, documentation, and other necessary files. It also creates input and output directories, and sets the working directory to /opt/src.
Once the Dockerfile is written following the three previously explained stages, it can be used to build the container. The final image for this module is approximately 300 MB. It’s important to note that the final output will be synchronized with the local host folder through Docker volumes, allowing for easy access to input and output data. For more information on how to build and run the container, please refer to the execution section.
INTERNAL WORKFLOW
The internal workflow of the module is executed when the user runs qlimr.py with specified arguments. In this context, qlimr.py acts as a wrapper
for the entire module’s workflow. It comprises five key components that work together to achieve the desired outcome. The main steps involved in
the workflow are described in the order of execution as follows:
Important
Create configuration file. The first step involves creating a configuration file that captures all the specified inputs provided by the user as arguments when running qlimr.py. This process generates a YAML file named
config.yamlinside the input folder, reflecting the data supplied by the user. Any flag arguments not provided will be left blank. Within this file, three main YAML keys categorize the input parameters: \(\texttt{input}\), \(\texttt{options}\), and \(\texttt{outputs}\).
Validate the input data. Once the configuration file has been created, the next step is to validate the input parameters provided by the user. The wrapper will execute a Python script named
Validator.py, which reads the config.yaml file generated in the previous step. This script checks the validity of the input data against a well-established specification outlined in a YAML file calledOpenAPI_Specifications_QLIMR.yaml. This specification defines the variable data types for all inputs and outputs. The validator compares the user-provided information against these specifications, ensuring that the correct data types (e.g., double or string) are used. If a data type mismatch occurs, the workflow will stop, requiring the user to provide a correct input value. If any input value is missing or falls outside the valid domain, default values will be used, as detailed in the input section. Upon completion of the validation process, a new file namedconfig_validated.yamlwill be created. This file will contain all the input parameters provided by the user, along with the default values filled in for any previously blank spaces in the config.yaml file. This validated file will be used by the C++ source code.
Pre-process the given EoS. The next step is to prepare the EoS data file provided by the user for use in the C++ source code. The wrapper will execute another Python script named
preprocess.py. This script first checks if a file is provided that adheres to the established conventions and formats (refer to the input section for more information). The naming convention for this file is alwayseosin either .csv or .h5 format. At the end of this step, a new validated file calledeos_validated.csvwill be created, containing only two columns: the first for total energy density \(\varepsilon\) and the second for pressure \(p\). If the user has already provided an EoS file with the same name and the correct two-column format, using a comma as the separator, this part of the workflow will be skipped.
Execute the C++ source code. The next step is to execute the most critical part of the workflow. The wrapper will run the C++ source code executable named
qlimr, which was created during the Dockerfile instructions. This executable computes all the quantities requested by the user, displays the results of the global observables on the screen, and creates two types of output files in the output folder. The first file,observables.csv, contains all the global quantities, while multiple files namedlocal_functions_#.csvare generated, each containing the local functions corresponding to different values of the central energy density. For detailed information about the implementation and structure of the source code, please refer to the source code subsection.
Post-process the output. The final step is to post-process the output data. The wrapper will execute a Python script called
postprocess.py, which converts observables.csv to HDF5 format if requested by the user through the input flag parameters.
To summarize the ideas presented in this subsection, below is a flowchart that represents the entire process of building the container and the workflow contained in the wrapper script.
Source code
This subsection explains the implementation of the source code, detailing the input parameter options available to the user and describing the outputs, including their column conventions.
IMPLEMENTATION
The primary goal of this module is to compute gravitational observables based on a barotropic equation of state (EoS). The functionality of the source code can be categorized into two cases depending on user needs:
Important
A. Sequence of Neutron Stars Calculation: The user wants to compute global quantities and local functions for an entire sequence specified by a range of central energy densities and a resolution in mass. The specified resolution means that the mass difference between adjacent data values in the sequence should be less than the provided value.
B. Single Star Calculation: The user wants to compute global quantities and local functions for a single neutron star, defined by a specified central energy density.
In both cases, the code allows users to select which observables they wish to compute and whether they want to obtain the local functions. To obtain the observables, it is necessary to solve the ordinary differential equations (ODEs) based on the EoS input data. The general approach is as follows:
Note
Read the Equation of State data file.
Interpolate the data imposing monotonicity.
Compute observables according to the specified case.
Export output quantities in the desired format.
To achieve this, the source code is organized into different header files (.hpp) with their corresponding .cpp files, each performing specific tasks. Descriptions of the main tasks associated with each file are provided below, presented in the same order as the execution of the source code. If users want detailed information about every method and function in each class contained in those files, they can refer to the source code itself, which is commented for that purpose.
A_Input : This is the first step in the calculation process. Its main goal is to read the user’s input, as specified by the provided parameters,
and to read the EoS file data, converting it into dimensionless values (see the units in one of the appendices).
B_Interpolation : After the Equation of State has been read and converted into dimensionless units, Steffen’s method is employed for interpolation.
This method guarantees monotonicity, enabling us to accurately compute \(p(h)\) and \(\varepsilon(h)\), which are essential for solving the TOV
equations in pseudo-enthalpy form. A key component of this part of the code is the initialize function, which is used to interpolate all relevant
ODE solutions throughout the code.
C_Zeroth_Order : This section of the code solves the TOV equations in pseudo-enthalpy form. It includes the integrator and sets the initial
conditions for the problem. The variables \(\texttt{NS_R}\) and \(\texttt{NS_M}\) represent the radius and mass of the neutron star, respectively.
These variables are computed after solving the ODE system by evaluating the solutions \(M(h)\) and \(R(h)\) at the surface when \(h=0\).
Upon completing this routine, the following local functions are obtained: \(p(R)\) , \(\varepsilon(R)\), \(M(R)\), and \(\nu(R)\).
D_First_Order : This section of the code uses the solutions of the TOV system to solve a second-order ODE, which enters at order
\(\mathcal{O}(\epsilon)\). It includes the integrator for this ODE and sets the initial conditions to obtain \(\varpi^{(1)}_{1}(R)\) and
\(\varpi'^{(1)}_{1}(R)\) as the solutions. Once these functions are obtained, the code computes the dimensionless moment of inertia, referred to as
\(\texttt{NS_Ibar}\).
E_Second_Order : This section of the code handles the equations that enter at order \(\mathcal{O}(\epsilon^{2})\). It includes two integrators
for solving \(h^{(2)}_{2}\) and \(k^{(2)}_{2}\) for the \(\ell=2\) mode: the homogeneous and inhomogeneous integrators, along with the initial conditions of the problem.
For the \(l=0\) mode, there is another integrator for solving the \(\xi^{(2)}_0\) and \(m^{(2)}_0\) system, as well as an integrator
for the function \(y\), which is used to obtain the tidal deformability.
After solving these systems, the code calculates the dimensionless tidal Love number \(\bar{\lambda}\), the dimensionless quadrupole moment,
\(\bar{Q}\) and the corrections to the radius \(\delta R_{\star}\) and mass \(\delta M_{\star}\). These quantities are stored in the
following variables: \(\texttt{NS_Lbar}\), \(\texttt{NS_Qbar}\), \(\texttt{NS_dR}\), and \(\texttt{NS_dM}\), respectively. By solving
all these equations, this part of the code computes the folllowing local functions: \(y(R)\), \(h^{(2)}_2(R)\), \(k^{(2)}_2(R)\), \(m^{(2)}_2(R)\),
\(\xi^{(2)}_2(R)\), \(m^{(0)}_2(R)\), \(\xi^{(0)}_2(R)\), and \(h^{(0)}_2(R)\).
F_Observables : This section of the code manages the application of previously defined methods based on user requests. Depending on the
combination of global quantities specified, a subroutine optimizes the calculation of the ODEs implemented earlier. For example, if the user
requests only the tidal Love number, there is no need to compute all the other second-order ODEs; instead, only the TOV system needs to be solved.
This optimization applies to all potential combinations of requests. Additionally, when the user selects Case A, this part of the code not only
handles the mass resolution but also outputs the local functions and the stable branch if requested. Thus, this section establishes the case type
and solves the problem in an optimized manner.
G_Output : This final section is responsible for exporting the outputs in the user-requested format, either .csv or .h5, while also displaying
the global quantities on the screen. Additionally, it generates a file named status.yaml, which contains a code indicating whether the entire
execution was successful. This code can be read by the calculation engine, providing a means to track any errors originating from the source code.
Attention
Internal Class Dependencies: The dependencies between classes are straightforward; each class in a header file relies on the classes defined in the preceding header files. This sequential dependency is crucial, as solving the equations at a higher order requires the results from the previous orders.
The following flowchart illustrates the overall design of the source code and summarizes its functionality. The left branch represents Case A, while the right branch represents Case B. The resolution algorithm for Case A is presented below the flowchart.
RESOLUTION ALGORITHM
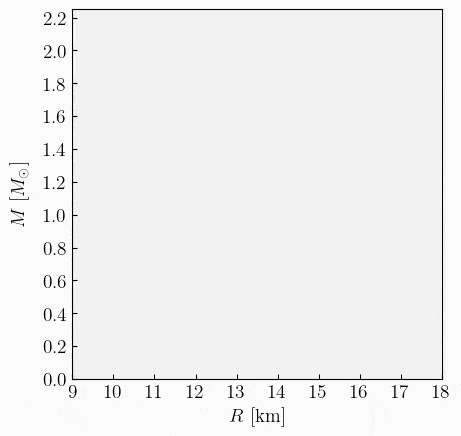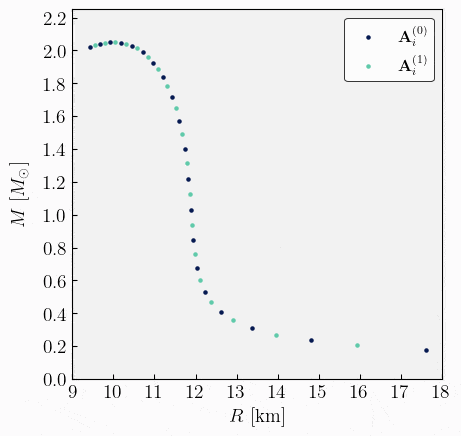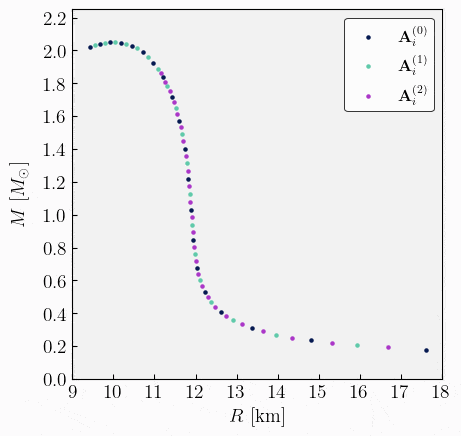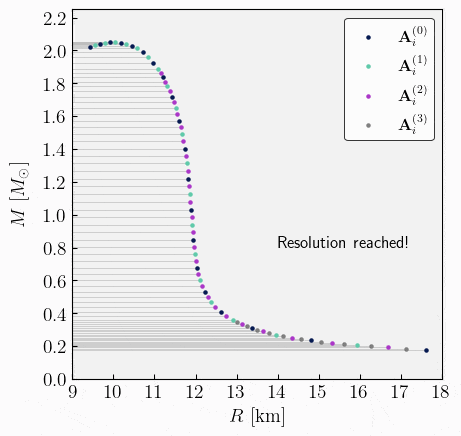
Create an initial set. The first step involves selecting a set of central energy densities to compute the neutron star (NS) sequence. The initial distribution in the source code is fixed at \(n=20\) values, arranged on a logarithmic scale. This distribution is determined based on the user’s specified initial and final central energy densities for the sequence.
The set of central energy densities can be represented mathematically as
\[\begin{equation} \boldsymbol{\varepsilon}^{(0)}_{c} = \{\varepsilon^{(0)}_{c,1}, \, \varepsilon^{(0)}_{c,2}, \, ..., \, \varepsilon^{(0)}_{c,n} \} \ . \end{equation}\]Here, the superscript \((0)\) indicates that this is the initial vector from which the sequence begins, while the subindex \(i\) in \(\varepsilon^{(0)}_{c,i}\) labels the different values in the set. Specifically, \(\varepsilon^{(0)}_{c,1}\) represents the initial value of central energy density specified by the user, and \(\varepsilon^{(0)}_{c,n}\) denotes the final value.
Get observables. Every initial condition specified by a central energy density yields a neutron star solution. The second step is to compute the gravitational observables requested by the user for each value in the previously defined set. We will assume the user is interested in all possible global quantities. These can be represented as follows,
\[\begin{split}\begin{align} \mathbf{A}^{(0)}_{1} &= \{ \varepsilon^{(0)}_{c,1}, \, R_{\star,1}^{(0)}, \, M_{\star,1}^{(0)}, \, \bar{I}^{(0)}_{1}, \, \bar{\lambda}^{(0)}_{1}, \, \bar{Q}^{(0)}_{1}, ...\} \ , \\[2ex] \mathbf{A}^{(0)}_{2} &= \{ \varepsilon^{(0)}_{c,2}, \, R_{\star,2}^{(0)}, \, M_{\star,2}^{(0)}, \, \bar{I}^{(0)}_{2}, \, \bar{\lambda}^{(0)}_{2}, \, \bar{Q}^{(0)}_{2}, ...\} \ , \\[1ex] &\vdots \\[1ex] \mathbf{A}^{(0)}_{n} &= \{ \varepsilon^{(0)}_{c,n}, \, R_{\star,n}^{(0)}, \, M_{\star,n}^{(0)}, \, \bar{I}^{(0)}_{n}, \, \bar{\lambda}^{(0)}_{n}, \, \bar{Q}^{(0)}_{n}, ...\} \ .\\[2ex] \end{align}\end{split}\]
Check resolution. Once we have computed all the observables from the initial set \(\boldsymbol{\varepsilon}^{(0)}_{c}\), we need to verify whether the resolution in mass is sufficient. Specifically, this means checking that the difference in mass between adjacent points on the mass-radius curve is less than the specified resolution provided by the user. If this condition is not met for any pair of points, we must add a new point between those values. This involves selecting a new central energy density that yields a mass value within the interval that does not meet the resolution criteria.
One possible way to create that new value is by taking the average of the central energy densities that correspond to the two masses being checked. This can be expressed mathematically as:
\[\begin{split}\begin{align} &\mathsf{If} \ \ \lvert M_{i+1} - M_{i} \rvert > \Delta M^{\textrm{(user)}} \\[2ex] &\mathsf{Create} \ \ \varepsilon^{(1)}_{c,i} = \dfrac{\varepsilon^{(0)}_{c,i+1} + \varepsilon^{(0)}_{c,i}}{2} \\[2ex] &\mathsf{Add \ to} \ \ \boldsymbol{\varepsilon}^{(1)}_{c} . \end{align}\end{split}\]Here, \(M_{i+1}\) is the mass obtained given the central energy density \(\varepsilon^{(0)}_{c,i+1}\), and \(M_{i}\) is the mass obtained from \(\varepsilon^{(0)}_{c,i}\). The algorithm must check the resolution in mass for every pair of adjacent points. Whenever it finds that the resolution is not met, it will create a new value of central energy density and add it to the new set \(\boldsymbol{\varepsilon}^{(1)}_{c}\). At the end, we obtain a new set of central energy densities given by
\[\begin{equation} \boldsymbol{\varepsilon}^{(1)}_{c} = \{\varepsilon^{(1)}_{c,1}, \, \varepsilon^{(1)}_{c,2}, \, ..., \, \varepsilon^{(1)}_{c,m} \} \ . \end{equation}\]
where \(m \leq n-1\).
Get observables. Since a new set of central energy densities has been generated, it can be used to compute all observables again. This will yield a new set in which additional points on the mass-radius curve are obtained to improve resolution. The new set can be expressed as follows,
\[\begin{split}\begin{align} \mathbf{A}^{(1)}_{1} &= \{ \varepsilon^{(1)}_{c,1}, \, R_{\star,1}^{(1)}, \, M_{\star,1}^{(1)}, \, \bar{I}^{(1)}_{1}, \, \bar{\lambda}^{(1)}_{1}, \, \bar{Q}^{(1)}_{1}, ...\} \ , \\[2ex] \mathbf{A}^{(1)}_{2} &= \{ \varepsilon^{(1)}_{c,2}, \, R_{\star,2}^{(1)}, \, M_{\star,2}^{(1)}, \, \bar{I}^{(1)}_{2}, \, \bar{\lambda}^{(1)}_{2}, \, \bar{Q}^{(1)}_{2}, ...\} \ , \\[1ex] &\vdots \\[1ex] \mathbf{A}^{(1)}_{m} &= \{ \varepsilon^{(1)}_{c,m}, \, R_{\star,m}^{(1)}, \, M_{\star,m}^{(1)}, \, \bar{I}^{(1)}_{m}, \, \bar{\lambda}^{(1)}_{m}, \, \bar{Q}^{(1)}_{m}, ...\} \ .\\[2ex] \end{align}\end{split}\]Stack and sort. Given the two sets of observables \(\mathbf{A}^{(0)}_{i}\) and \(\mathbf{A}^{(1)}_{i}\), the next step is to combine the two sets into one and sort the data in order of increasing energy density. This process will result in a new sorted set that looks like,
\[\begin{split}\begin{align} \mathbf{A}^{(0)}_{1} &= \{ \varepsilon^{(0)}_{c,1}, \, R_{\star,1}^{(0)}, \, M_{\star,1}^{(0)}, \, \bar{I}^{(0)}_{1}, \, \bar{\lambda}^{(0)}_{1}, \, \bar{Q}^{(0)}_{1}, ...\} \ , \\[2ex] \mathbf{A}^{(1)}_{1} &= \{ \varepsilon^{(1)}_{c,1}, \, R_{\star,1}^{(1)}, \, M_{\star,1}^{(1)}, \, \bar{I}^{(1)}_{1}, \, \bar{\lambda}^{(1)}_{1}, \, \bar{Q}^{(1)}_{1}, ...\} \ , \\[2ex] \mathbf{A}^{(0)}_{2} &= \{ \varepsilon^{(0)}_{c,2}, \, R_{\star,2}^{(0)}, \, M_{\star,2}^{(0)}, \, \bar{I}^{(0)}_{2}, \, \bar{\lambda}^{(0)}_{2}, \, \bar{Q}^{(0)}_{2}, ...\} \ , \\[1ex] &\vdots \\[1ex] \mathbf{A}^{(1)}_{m} &= \{ \varepsilon^{(1)}_{c,m}, \, R_{\star,m}^{(1)}, \, M_{\star,m}^{(1)}, \, \bar{I}^{(1)}_{m}, \, \bar{\lambda}^{(1)}_{m}, \, \bar{Q}^{(1)}_{m}, ...\} \ ,\\[2ex] \mathbf{A}^{(0)}_{n} &= \{ \varepsilon^{(0)}_{c,n}, \, R_{\star,n}^{(0)}, \, M_{\star,n}^{(0)}, \, \bar{I}^{(0)}_{n}, \, \bar{\lambda}^{(0)}_{n}, \, \bar{Q}^{(0)}_{n}, ...\} \ .\\[2ex] \end{align}\end{split}\]Repeat. From the newly stacked and sorted set, the algorithm can repeat steps 3 to 5 until the desired resolution is achieved. The entire algorithm is illustrated in the following GIF image